Apis species comparison: Gut Microbiota and Host Evolution
Honeybees serve as a powerful model for studying gut microbiota evolution in the context of host ecology and evolutionary history. Unlike the highly diverse and variable microbiomes of humans, primates, or mice, honeybees harbor a small, conserved set of gut microbes shaped by well-documented ecological and evolutionary forces. This makes them a tractable system for understanding how gut microbiota are distributed and evolve across hosts.
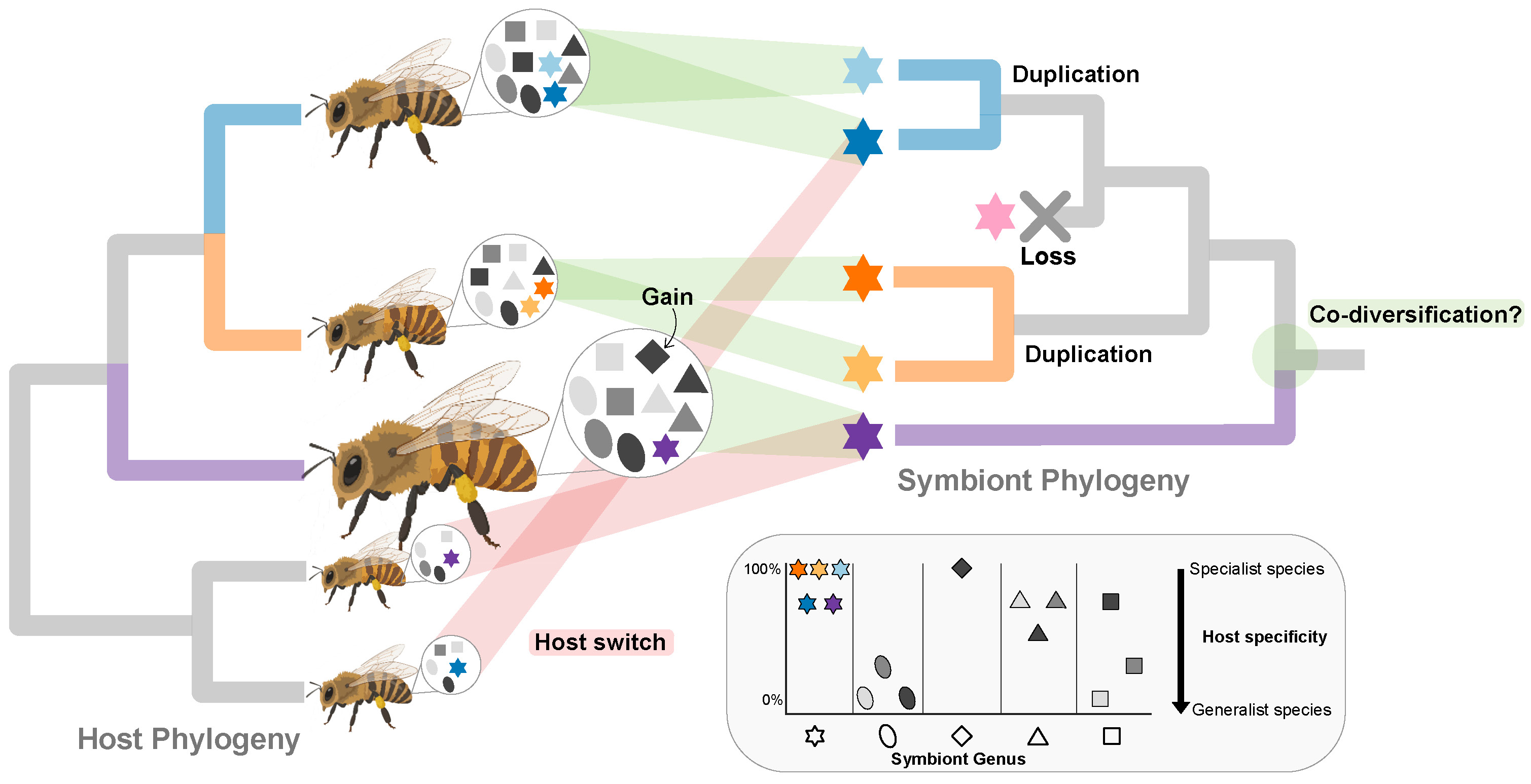
This research is essential for advancing our understanding of microbiome interactions across species, but has been limited by a lack of high-resolution genomic data. To address this, we performed deep shotgun metagenomic sequencing on 200 honeybee workers across five Apis species, sampled from Malaysia and India.
From this dataset, we recovered thousands of metagenome-assembled genomes (MAGs), including several previously uncharacterized bacterial species. We observed both host-specialist and host-generalist bacterial lineages, with some generalists appearing host-specific only at the strain level—suggesting recent host switching events.
We found no evidence of codiversification between microbes and hosts. Instead, our results support a model of dynamic community turnover driven by symbiont gain, loss, and replacement. These changes were also reflected in functional profiles—for example, variation in genes involved in degrading pollen-derived pectin.
Key Objectives
- Investigate how gut microbiota composition and function vary across closely related host species
- Resolve evolutionary patterns beyond species-level taxonomies
- Identify differences in functional traits, symbiont gain/loss across host species
Methodology & Highlights
- Generated hundred of metagenome-assembled genomes (MAGs), including several previously uncharacterized microbial species
- Identified host-generalist and host-specialist bacterial lineages, with strain-level divergence suggesting recent host switching
- Found functional differences among species, such as pectin degradation in certain hosts’ microbiomes pointing to host-specific functions
- Found no signal of co-diversification between hosts and microbes; instead, dynamic symbiont loss, gains and switches over evolutionary timescales shaped gut microbiota evolution
Publication
This study has been released as a preprint on bioRxiv and is currently under peer review.