Priority Effects in the honeybee gut
Gut microbial communities often differ at the strain level even among closely related individuals, but the ecological mechanisms driving this variation are not well understood. One potential driver is priority effects—differences in the timing or order of microbial colonization—which can lead to lasting differences in community structure even under similar environmental conditions.
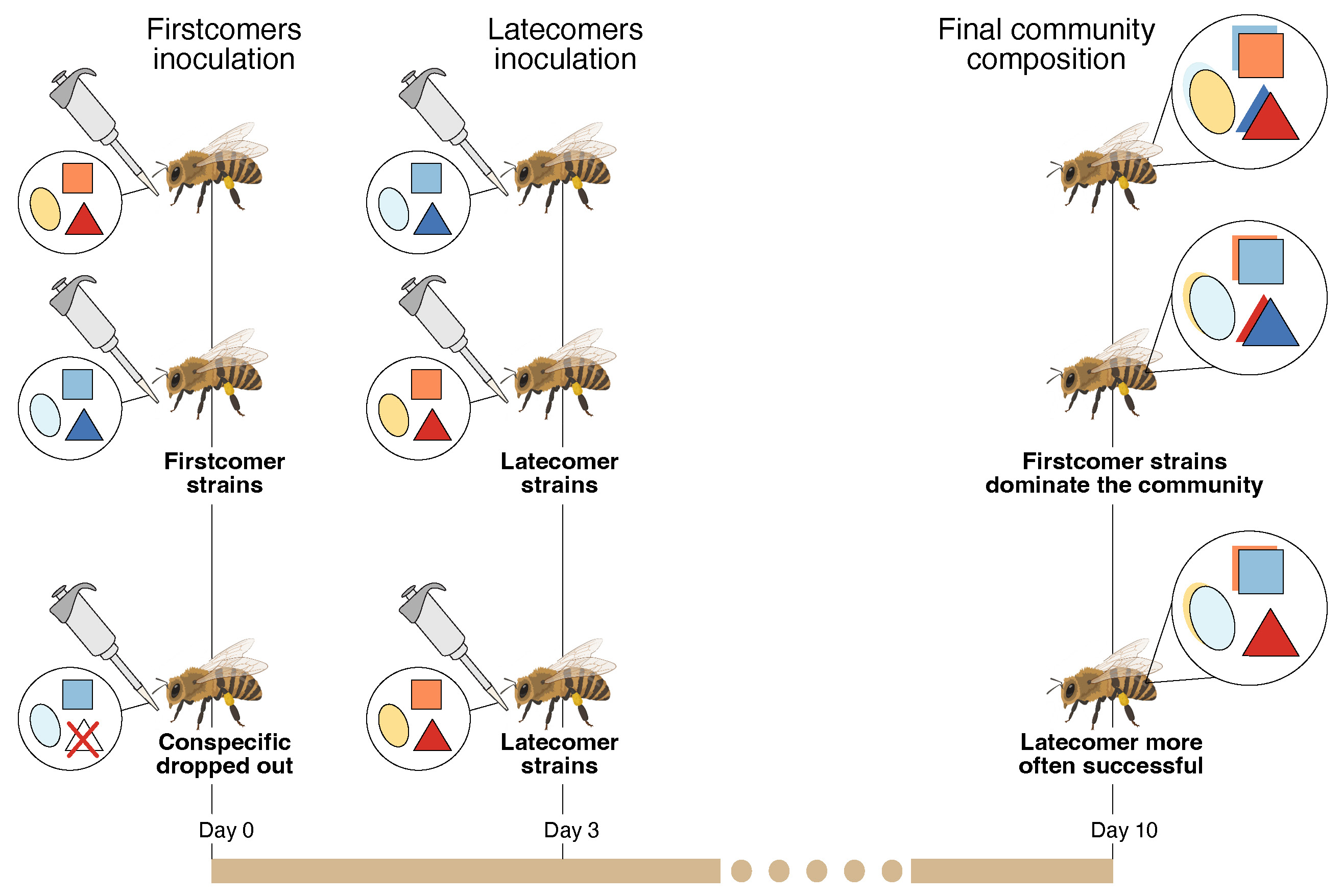
In this project, we test the role of priority effects in shaping gut microbiota using honeybees as an in vivo model. Honeybees host a simple, well-characterized gut microbiome, and their colonies offer a tractable system for studying strain-level dynamics under controlled conditions.
We sequentially colonized microbiota-depleted honeybees with two synthetic communities composed of the same 12 bacterial species, but with different strain variants. Across replicates, we observed that firstcomer strains consistently dominated, though the strength of this effect varied by strain and species. Removing select strains from the first wave only partially restored the ability of latecomer conspecifics to establish—suggesting that priority effects also occur across species boundaries.
These results highlight the importance of colonization order in shaping microbial communities at the strain level and offer insights into the stability and variability of host-associated microbiota.
Context
- Understanding strain-level assembly rules is essential for predicting and designing microbiomes
- Most studies have not probed microbe-microbe interactions within species
- Honeybees allow high-resolution, reproducible studies in a natural but tractable host system
Experimental Design & Technical Challenges
- Designed a controlled colonization experiment using synthetic communities of Lactobacillus and Bifidobacterium strains
- Developed a high-throughput compatible PacBio metagenomic approach to sensitively detect and quantify individual strains in vivo
- Implemented multiple combinations and replicates to evaluate robustness and generality of observed priority effects
Findings & Implications
- Pilot results show species-level coexistence through niche partitioning, consistent with previous work in honeybees
- At the strain level, strong priority effects were observed—early-arriving strains dominated over conspecifics introduced later
- Unique strain of Lactobacillus showed weak priority effects, making it an interesting candidate for development of robust probiotics
- Interactions within and between close Bifidobacterium species shape priority effects but not between genera, emphasising the importance niche partitioning
Publication
Manuscript in preparation.